| Algorithmic "Oritatami" Self-Assembly Lab |  |
Oritatami is a mathematical model of a phonomenon called cotranscriptional folding of RNA, aiming at widening applications of RNA origami, one of the most significant experimental breakthroughs in molecular engineering and understanding the nature of cotranscriptional folding, the way that cells control RNA folding in vivo. This model is a novel topic of algorithmic self-assembly, which quests for efficient designs of self-assembly systems theoretically and experimentally (see Self-assembly.net).
RNA origami, proposed by Geary, Rothemund, and Andersen in Science, is an experimental architecture of nanoscale RNA rectangular tiles that self-assemble uniquely from an RNA sequence being folded cotranscriptionally.
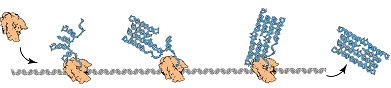
An RNA transcript starts folding upon itself to take the most stable conformation, while it is still being synthesized, in a continous process controlled by the rate of strand synthesis, instead of remaining sequential until it will have been fully transcribed. This way of folding is called cotranscriptional or kinetic folding. In the cotranscriptional folding, locally-stable structures of RNA will be preferred over some folds with higher stability because they would require first unfolding parts of the strand in order to form.
Theoretical models for programming self-assembly by cotranscriptional folding are needed, as Winfree's abstract tile assembly model (aTAM) and its variants have yielded tremendous successes in experimental DNA tile self-assembly, which self-assemble a structure by letting unit DNA tiles attach to each other in some pre-programmed manner.
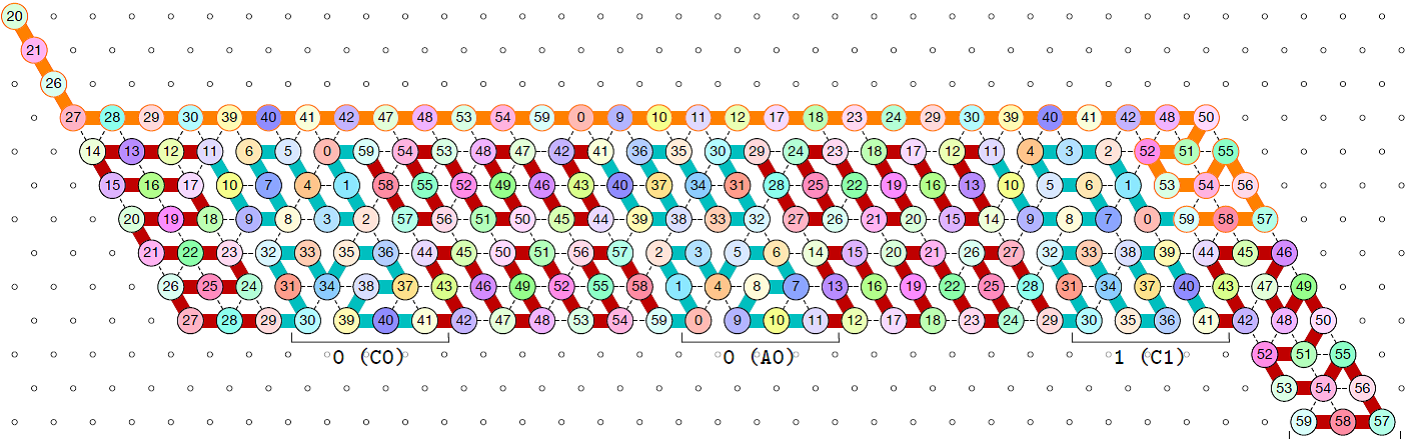
Oritatami is NOT for predicting most likely conformations of RNA. It rather enables us to implement computational devices out of RNA that take advantage of sequential folding to do something practical, such as count. With Geary, Meunier, and Schabanel, for instance, we propose an oritatami binary counter, which suggests a way to use cotranscriptional folding for biomolecules to count in vivo.
News
| The website of DLT2018 open!! | |
| TAF decided to support DLT2018. | |
| The 1st International Workshop on oritatami systems (Oritatami2017) was held in Fayetteville, AR, USA. | |
| An invited article on oritatami system was published in SIAM News | |
| NICT International Exchange Program decided to support DLT2018, which we will host in summer 2018. | |
| The paper "Rule set desing problems for oritatami system" by the alumnus Makoto Ota was accepted | |
| CIAA 2016 Best Paper Award (Sheng Yu Award) winner | |
| Our collaborator Nicolas Schabanel was invited to give a talk about the oritatami system at AUTOMATA 2016 (Zurich, Switzerland, June 15-17, 2016) | |
| JSPS Grant-in-Aid for Young Scientists (A) awarded. |
 |
 |
 |
 |
 |